Cell Biology 05: The Cytoskeleton Part I: Actin
These are notes from lecture 5 of Harvard Extension’s Cell Biology course.
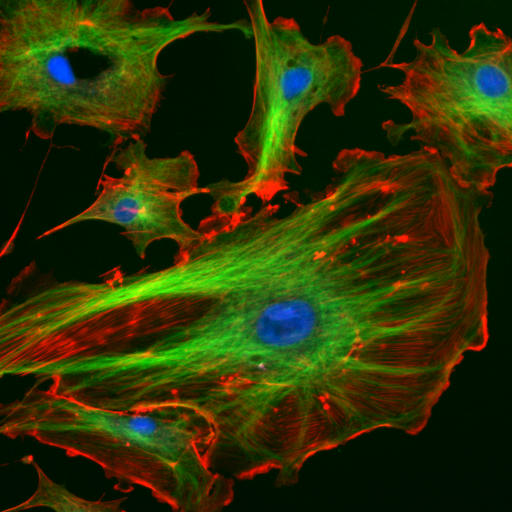
The cytoskeleton is many things to the cell: a structural scaffold giving the cell shape, an intracellular transport system, a driver of cell motility, and a mediator of cell division, to name a few of the most important. Accordingly the cytoskeleton gets two lectures, devoted to its two most important pieces: this week on actin (red below) and next week on microtubules (green below; the blue are nuclei).
The cytoskeleton is a dynamic 3D protein network connected to the membrane and some organelles. There are three elements to the cytoskeleton, each a chain of specific protein units:
- microfilaments made of actin monomers
- microtubules made of tubulin dimers, an
- intermediate filaments made of various other proteins.
Because the cytoskeleton is modular, consisting of polymers of individual proteins, it can expand or contract to change cell shape, move and respond to stimuli. Cytoskeletal systems vary greatly across cell types, in part responding to signals received from outside the cell.
Today’s lecture will focus on microfilaments but briefly here is a brief introduction to the other two things. Microtubules extend throughout the cell, providing an organizational framework for organelles. They give shape to cilia and flagella in eukaryotes, and they form the spindles that pull sister chromatids apart during mitosis. Intermediate filaments are tissue-specific. They support the nucleus and nuclear membrane, provide cell structural support and promote barrier formation in hair, skin and nail cells. To our knowledge, they have no transport role – i.e. they are not substrates for motor proteins to walk upon. By contrast, microfilaments and microtubules both serve as transport systems. Kinesin and dynein, powered by ATP, walk on microtubules (anterograde and retrograde respectively), as will be covered in more detail next lecture. Myosin walks on actin microfilaments, the focus of today’s lecture.
Microfilaments are polymers of actin. They can form complex structures by being held together in different ways by a dizzying variety of actin-binding proteins. Microfilaments organize the plasma membrane: animal cells have a ‘cell cortex‘ which is an actin-rich layer just under the membrane. Actin also forms the contractile ring which cinches the cell into two during mitosis.
Actin microfilament formation is portrayed in the Inner Life of the Cell video, although it is perhaps my least favorite part of this really awesome video. Susan Lindquist pointed out that this video makes two major sacrifices in order to make what’s going on in the cell visually intelligible: (1) concentration and (2) speed. Unlike the elegant negative space of the video, all the molecules in a cell are packed in right next to each other, sliding past one another and interacting constantly. And molecules at body temperature move at about 25 miles per hour, while the cell is only 10-30 μm across [wikibooks], meaning that every passing second provides an astronomical number of opportunities for molecular interactions. These two factors – speed and density – are crucial for everything that happens in the cell, and omitting them is a necessary sacrifice for any video depiction, but to me it is especially problematic in showing actin formation. This video makes it look as though the actin proteins just know to come together by magic to form filaments:
Actin microfilaments also form lamellipodia and filapodia, two structures critical for the forward mobility of motile cells. Lamellipodia are a mesh-like network of actin filaments inside the cytoplasm that extend a smooth leading edge at the ‘front’ of migrating cells. Filapodia are finger-like, spiky protrusions of actin filaments that extend more of a jagged edge. Both are required for proper cell migration in some cells, and both are composed of actin – just with different organizing proteins giving rise to different superstructures of microfilaments. Actin is also a structural component of microvilli, another feature seen only in some cell types (intestines and ova), and stress fibers important for muscle contraction, and adherens junctions that link cells together (epithelial cells and cardiomyocytes).
Actin comes in three main isoforms (and a few minor ones we won’t cover here). The big three are predictably named alpha-actin, beta-actin and gamma-actin and coded for by the three genes ACTA1, ACTB and ACTG1. Each isoform is ~375 amino acids, only ~25 of which vary between these three isoforms – and only 20% of which vary across species from algae to humans, indicating strong conservation. But that little bit of variability gives rise to great differences in function between isoforms. Alpha is important in contractile structures such as the contractile ring for mitosis; beta is important in the cell cortex and at the leading edge of motile cells (lamellipodia and filapodia) and gamma is important in stress fibers. Collectively these make up 10% of cell mass in muscle cells [according to class; 20% according to Wikipedia].
Regardless of isoform (alpha/beta/gamma), actin is called G-actin (for globular) when it’s a monomer and F-actin (for filamentous) when it’s a polymer. Actin monomers are polar with a ‘barbed’ (+) end and a ‘pointed’ (-) end. In the spokes of the cytoskeleton, the (+) end points to the cell surface and the (-) points to the center of the cell. The (-) end has an ATP binding cleft, and actin must bind either ADP or ATP in order to polymerize at all, with much higher inclination towards polymerization when ATP-bound than when ADP-bound. Actins usually bind an ion as well (most often Mg2+, sometimes K+ or Na+) and ion concentrations determine whether filament formation or dissociation is favored.
Filament formation is said to take place in three steps:
- Nucleation. This is the rate limiting step – actin monomers have limited affinity to form oligomers with one another but higher affinity to join an existing filament. It’s like how your first million is your hardest to make, or how no one wants to come to the party until they know who else will be there. Specifically, actin dimers dissociate very easily, but once a trimer forms, it is relatively stable and can act as a seed to attract other G-actin molecules. Apparently it takes three to tango.
- Elongation. Once nucleation has happened, and provided that ion and G-actin concentrations favor it, the filament will elongate rapidly. G-actin monomers prefer to join the (+) end of a filament, so that end will grow faster, but in many conditions the (-) end will grow as well.
- Steady state. Eventually the filament will grow to where the ambient concentrations of G-actin and requisite ions have been depleted enough that the rate of actin monomer dissociation from the filament equals the rate of elongation. The dissociation happens more at the (-) end and elongation more at the (+) end, so even in equilibrium, the filament is still dynamic and has a ‘treadmilling’ appearance.
The above is a slight simplification because filament nucleation and the equilibrium between elongation and dissociation also depend upon actin-binding proteins. Profilins (genes: PFN1-4) are small proteins that promote swapping out ADP for ATP in the actin cleft: they bind to the (+) end of ADP-bound G-actin when ATP is more abundant and release the ADP, allowing a swap to occur. Profilin also blocks (-) end growth, thus further promoting ‘treadmilling’. Cofilins (genes: CFL1, CFL2, DSTN) twist and destabilize filaments, promoting dissociation. Thymosin beta 4 (gene: TMSB4X) is another small protein that binds ATP-bound actin and physically blocks addition to the filament at either end – thus it prevents excessive elongation of filaments in conditions of abundant ATP.
Other proteins can completely ‘cap’ the ends of a filament, preventing assembly or disassembly. CapZ (genes: CAPZA1-3, CAPZB) are (+) end specific, tropomodulins (genes: TMOD1-4) are (-) end specific and are found in non-motile cells that, since they don’t move, want stable, non-expanding non-contracting actin filaments, and gelsolin (gene: GSN) is (+) end specific and has a Ca2+-dependent ability to sever filaments.
Nucleation is promoted by ‘actin nucleating proteins’ that help bring monomers together. Chiefly, formins (many genes) help to form long, straight filaments and the dimer Arp2/3 (genes: ARPC2 & ARPC3) helps to form branched filaments, for instance in lamellipodia.
Formins have two domains (called FH1 and FH2 for formin homology 1 & 2) that form a donut-shaped complex that promotes elongation at the (+) end. Specifically, FH1 is rich in proline, and profilin (see above) is attracted to proline-rich domains. So formin hangs out at the (+) end of F-actin, and recruits profilin which attracts ATP-bound G-actin, which can then be added to the filament. Formin thus effectively acts to increase the local concentration of G-actin above the critical threshold for elongation to occur. It also prevents (+) end capping.
Formin itself is regulated by Rho G-proteins. Remember, G-protein means GTPase, a protein that can bind GTP and hydrolyze it to GDP as an energy source. Certain extracellular signals can activate Rho-GEF (any of a bunch of different proteins with one shared domain) to load Rho with GTP. When (and only when) Rho is GTP-bound, it ‘opens’ formin into the donut configuration so it can do its job.
The Arp in Arp2/3 stands for actin-related protein. To do its job of promoting filament branching, it needs NPF (nucleation promoting factor) as well as WASp (Wiskott-Aldrich Syndrome protein). When Arp and WASp both bind to the side of a filament (i.e. not at the (+) or (-) end), they allow a new filament to form off to that side, with its (-) end pointed into the trunk of the original filament. This process is also regulated by a Rho protein, Cdc42 (gene: CDC42) which (when bound to GTP) regulates WASp which in turn activates Arp2/3.
A genus of bacteria called Listeria have figured out how to hijack our actin systems for their own benefit. Listeria has its own actin protein, ActA, which uses your own cells’ Arp2/3 to recruit your own actin monomers to form a ‘comet’ that propels it around the cell, as shown in this video:
To study actin, researchers often use:
- phalloidin from Amanita death caps, which binds to actin filaments (and stabilizes them, preventing depolymerization) – that’s both why it’s toxic and why it’s useful for imaging to see where actin is
- latrunculin from sponges, which binds G-actin to prevent filament formation, blocking cell motility and cytokinesis
- cytochalasin D from molds, which caps the (+) end of filaments, preventing elongation
- jaspakinolide from sponges [see Bubb 2000] which promotes nucleation by stabilizing actin dimers, making it more likely they’ll stay together long enough to form a trimer at which point they can start elongating into a filament. By doing so, it effectively lowers the critical concentration of G-actin required for filament formation.
Of course, the actin filaments are themselves organized into various superstructures. Cross-linking proteins help to determine what structure will form. In addition to Arp2/3, which as mentioned earlier promotes branching, also of note are the following:
- fimbrin binds two close-together filaments with the same polarity (i.e. ‘pointed’ in the same direction), important for forming those jagged spikes in filapodia.
- alpha-actinin (gene: ACTN1), a larger protein, can bind two filaments that are further apart than fimbrin.
- filamin (genes: FLNA, FLNB, FLNC) is a sort of ‘hinge’, binding two actin filaments at a large angle and thus helping to form a mesh of actin filaments.
- the ERM proteins (ezrin, radixin and moesin) link parallel actin filaments, with their (+) ends all pointed outward into the tips of microvilli.
- spectrin is a tetramer formed of 2 alpha (genes: SPTA1, SPTAN1) and 2 beta (genes: SPTB, SPTBN1,2,4,5) subunits pointed in opposite directions. The spectrin complex binds one actin filament in each end and is a critical part of forming the triangles that make up hexagons in the cell cortex (see below).
(thanks to Wikimedia Commons user kupirijo)
This spectrin tetramer is depicted in Inner Life of the Cell:
So far we’ve talked about the structural role of the microfilaments. They also have a transport role, serving as sort of a set of train tracks for ATP-powered motor proteins to move along. Myosin is the main motor protein that operates on microfilaments. It has a ‘head’ which both binds actin and binds ATP, a ‘neck’ which acts as a lever – its length determines the stride at which myosin walks – and a ‘tail’ which binds cargo to be transported. The tail is highly variable, as its sequence determines what cargo will be bound. The head must burn one ATP molecule for every ‘step’ it takes. Myosins are a huge family of proteins that come in 13 different classes (genes starting with MYO, MYH and MYL – the H and L are for heavy and light chain components). Here are just a few of the classes:
- Class I myosins couple actin filaments to the cell membrane and are involved in endocytosis. They have just one ‘head’ that binds actin and ATP.
- Class II do muscle contraction. They have two ‘heads’ allowing them to ‘walk’ by alternating which head binds.
- Class V are two-headed and ‘walk’ toward the (+) end of filaments, but they seem to spend most of their time just walking on the treadmill: they don’t transport vesicles so much as just keep them in the cell periphery. In yeast it organizes the organelles by virtue of its long neck region and 6 light chains and tails. But in animals, microtubules (topic for next week) deal with organelle organization.
- Class VI are two-headed and are the class that can ‘walk’ toward the (-) end, performing the retrograde movement of taking endocytic vesicles deeper into the cell.
People have studied myosin movements in vitro via a filament sliding assay: tether myosin to a glass side and then add ATP and fluorescently labeled actin filaments and see what direction the filaments move. These are the experiments that revealed that the majority of myosins (all but Class VI) move toward the (+) end, which you can see because the myosins push the (-) end outward when the myosin is tethered and the filament free-moving.
When you look at muscle fibers under a microscope you see two main parts: thick filaments, which are myosin II, and thin filaments which are actin. When motor neurons fire onto the muscle cells, post-synaptic pathways signal for the sarcoplasmic reticulum (a special word for the ER in muscle cells) to release its reserves of Ca2+. Myosin II is Ca2+-dependent, so when calcium rushes into the cytosol, myosin II starts burning ATP to try to ‘climb’ toward the (+) end of actin, but since it’s doing this from both ends, it doesn’t actually move, but instead pulls the actin filaments closer together. That’s what muscle contraction is at the molecular level. Here it is in video form:
And a bit more zoomed out:
Cell motility is important for finding food and avoiding prey in single-celled organisms, and for embryonic development, wound closure and infection/disease control in multicellular organisms. To move, cells have to have a ‘front’ and a ‘back’. At the front, actin filaments extend the cell forward via extending lamellipodia and filapodia. These cells also have a sort of ‘bottom’ where membrane proteins such as integrin adhere to the extracellular matrix (or a glass slide, etc. in the lab) relative to which the movement is occurring. At the ‘back’ of the cell, myosin II / stress fiber contraction pulls the cell away from old adhesion sites – often the adhesion proteins snap, with tails still bound to the old adhesion surface and the heads endocytosed into the cell. At the front and back, adapter proteins (Arp2/3 and formin respectively) join the actin network to the plasma membrane via membrane-anchored G proteins: at the front, Arp2/3 binds to membrane-anchored G proteins. Extracellular signals often induce the cell to move by signaling through G proteins, chiefly the Rho family proteins including Rac, Cdc42, GDI (GDP displacement/dissociation inhibitor).
discussion section
Suraneni 2012 studied mouse fibroblasts homozygous for ARPC3 knockout, thus having no functional Arp2/3 complex. The cells were unable to form lamellipodia. They still formed something that looked an awful lot like filopodia but which Suraneni did not characterize at the molecular level. The cells still moved around, but lacked directionality – they moved more like a random walk, and when a gradient of epidermal growth factor (EGF) was added they moved faster but failed to move up the gradient as they are supposed to.