Gene therapy immunogenicity primer
Jenn Abelin of Broad’s Proteomics Platform visited our lab to give us a primer on immunogenicity in gene therapy development. Nir Hacohen of Broad/MGH joined for the discussion. Here are my notes.
In gene therapies, your hope is to bring a new piece of DNA into a patient’s cells in order to express a gene that they need, or in order to perform gene silencing or other functions. The novel protein that you are causing their cells to produce has the potential to be immunogenic. How can one design the therapeutic transgenes to minimize this immune response? The purpose of today’s primer is to help us understand what pathways could lead to immune response and how one can design therapies to minimize this response.
Jenn Abelin | Background on the importance of HLA peptide presentation and understanding the immunogenicity of proteins
Jenn’s career has been dedicated to using mass spec to sequence HLA-bound peptides &mash; first at Broad, then Neon Therapeutics (later acquired by BioNTech), then Broad again. Most applications of this technology are in oncology but there are also people using this to de-risk biologics.
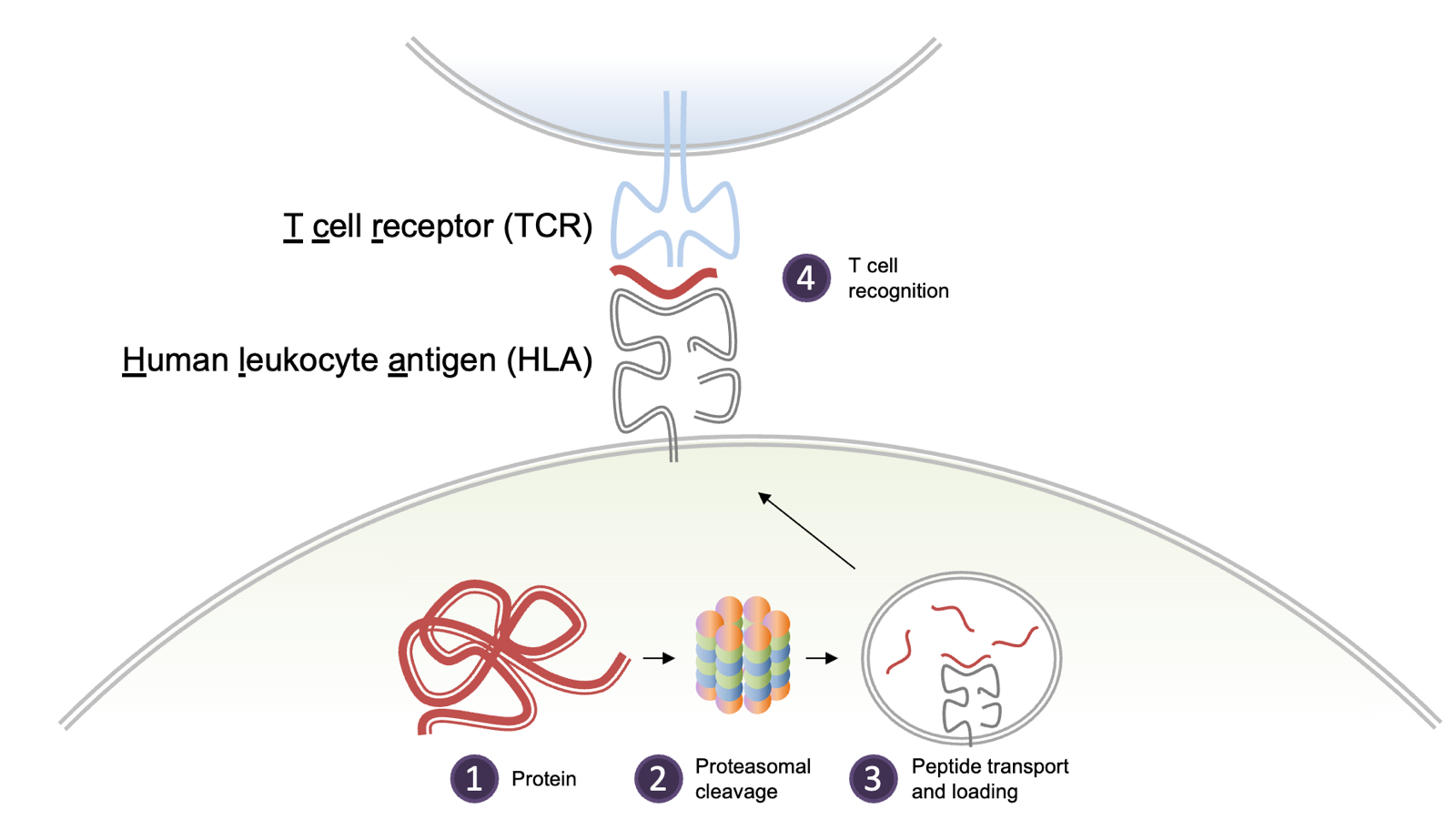
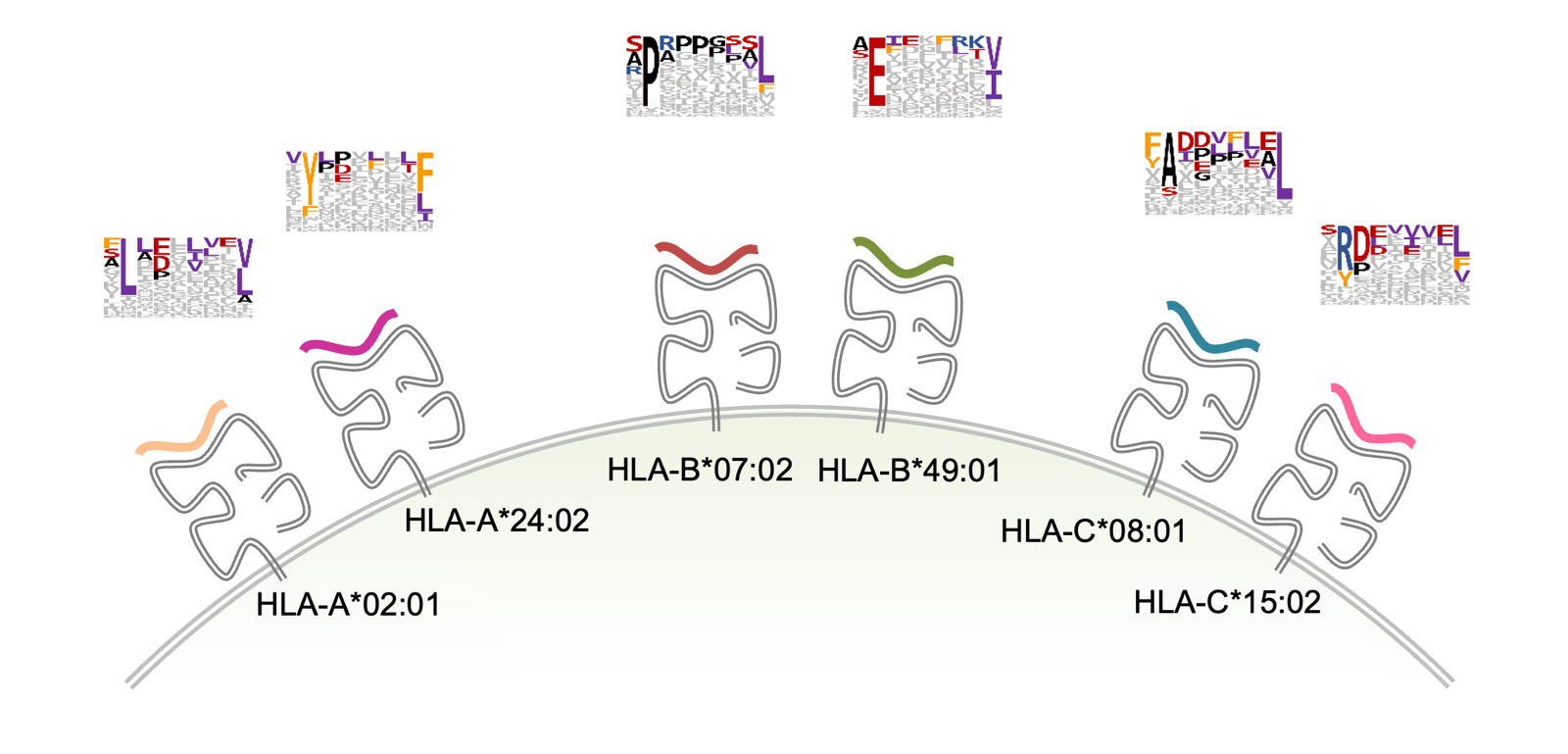
HLA Class I mediates cytotoxic T cell response, used to fight malignant or infected cells. HLA class I is a cell surface complex expressed on almost all nucleated cells in the body. The complex presents pieces of peptides from endogenous proteins naturally being degraded. Pieces of almost every protein in your body can be found there. The peptides comes from the proteasome but there are also special pieces of machinery that trim the peptides to just the right size to fit in the deep groove where they will be displayed. This system allows T cells to survey the health of cells throughout the body. The peptides have a half-life where some might stay for hours, other for days. HLA I is the most polymorphic region in the genome, with over 20,000 alleles in the population. There are a variable number of copies in tandem, so people have 3-6 different alleles. Each individual allele has a different motif that it likes to present. Class I HLA presents short peptides, usually 8-12 amino acids, and one allele of HLA I for instance, HLA-B*07:02, likes a P at position 2 and a L at the final position. HLA-A*24:02 prefers a Y and an F in those positions. And so on.
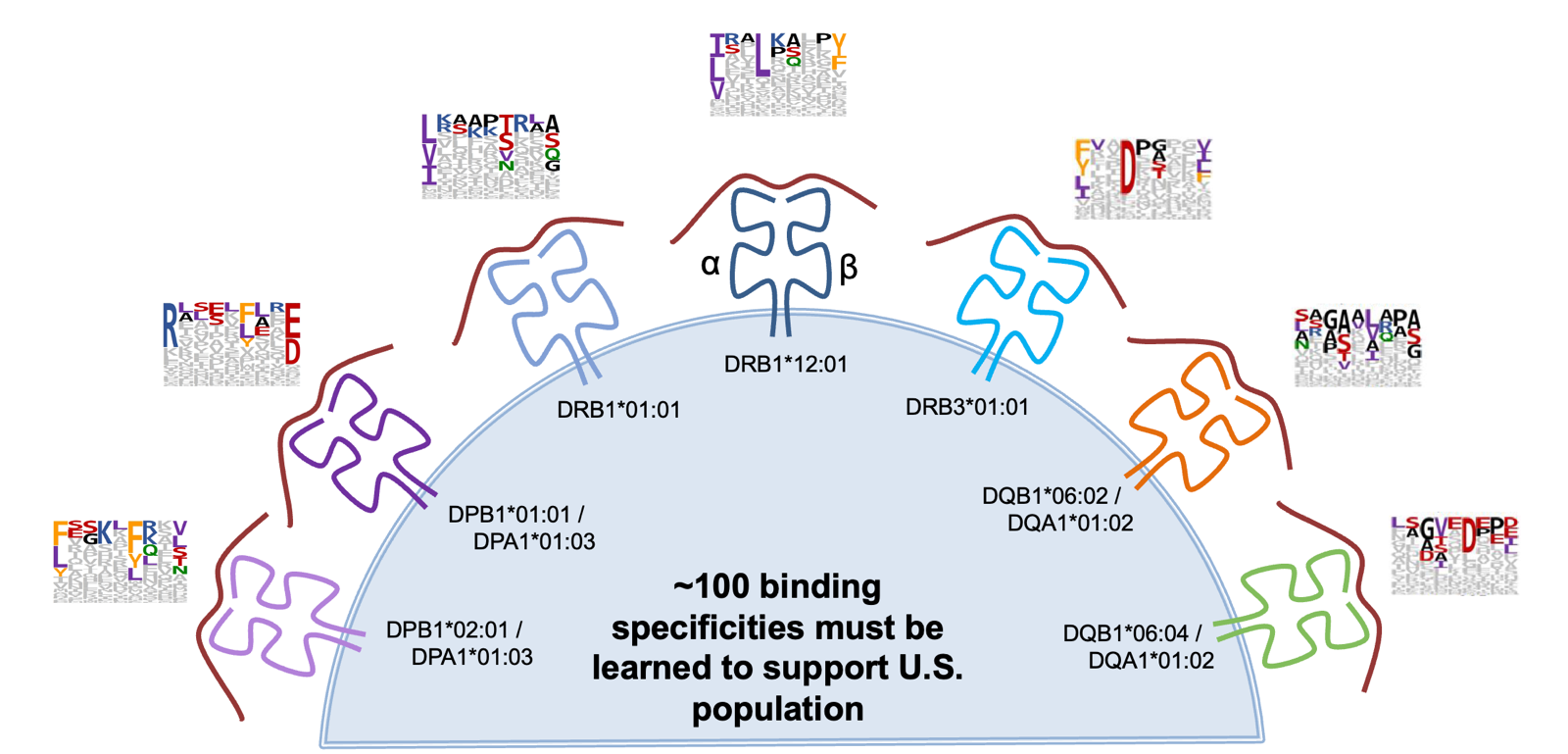
HLA Class II mediates B cell responses. Class II presents peptides to a different population of T cells, and is restricted only to antigen-presenitng cells like dendritic, macrophage, some endothelial. For immunogenicity of biologics, HLA II is the really interesting one. It can present not only endogenous proteins but also exogenous ones that are taken up via endocytosis. HLA II also has many different alleles, and people often have 12 different functional complexes due to combinatorics of how your DR, DP, DQ and A and B interact. In the endosome there is a placeholder protein called CLIP derived from CD74. Nascent peptides have to outcompete it and get trimmed back. The rules of what gets displayed are super complex and difficult to learn through sequencing the peptides that we see displayed.
A lot of people want to learn to predict which peptides will be displayed. This has resulted in better and better methods being invented:
- Biochemical pMHC binding assay. In the 1990s, people would use low throughput biochemical methods where you have to pick an allele and study it in a simplified system that loses a lot of hte original biology.
- Multi-allelic mass spectrometry. People realized you could isolate HLA I and HLA II molecules, elute the peptides off of them and sequence them with a mass spectrometer. This is high throughput and it’s unbiased, but, you only know that the peptide bound to any HLA on your cell line, you don’t know which HLA allele it bound to.
- Mono-allelic mass spectrometry. You take HLA-null cell lines, put in just 1 HLA allele, and then sequence the peptides bound to it. If you learn the rules for the top 100 HLA molecules, you can more or less predict immunogenicity across all 20,000 alleles.
- Immunoaffinity enrichment followed by mass spectrometry. Use antibodies to isolate the HLAs of interest; these antibodies are not perfect but some have decent specificity. You can also put affinity tags on the HLAs and then pull them down.
A lot of companies will now use these methods to pick the least immunogenic drug candidates [Karle 2020, Barra 2020]. They take multiple biologic drug candidates, put them on HLA-presenting cells, isolate the HLA II and sequence the peptides. They prioritize the candidates that present less. DR is expressed the highest. So they usually select 10 donors with diverse DR alleles and run the assay only on them. This is usually done for systemically delivered antibodies or extracellular protein therapeutics, where you are only worried about Class II responses, i.e. anti-drug antibodies (ADA). There is for example a Genentech poster showing that presentation in this assay is correlated with the incidence of ADA. Some people will use PBMCs since they express both Class I and Class II, and in oncology you can get them matched from the same patients you are hoping to treat. But in oncology people are working on the opposite problem — they want immune responses and are trying to find the most immunogenic peptides.
A ton of people are working on computational prediction of which peptides will be displayed [Abelin 2019]. Predictions are not perfect but are now pretty good and can guide which experiments you do. If you have limited resources, use computational methods to predict which HLA alleles are potentially most problematic for your biologic/protein sequence, then test only those alleles in the lab. The predictors only predict whether a peptide would get processed and presented though, not how immunogenic that peptide is. For example, they don’t at all look at whether the peptides are matched to endogenous host peptides expected to be non-immunogenic.
NetMHC is the most widely used prediction tool by industry. The Hacohen group has been working on hlathena (also available on Terra), as an alternative. Running both is worthwhile. hlathena captures the top 95 HLA class I alleles, and takes into account expression level. It’s true that lower expression means less likely immunogenicity.
Discussion
For systemically delivered gene therapies for the CNS, you mostly need to worry about HLA class I, since it is presented on neurons which are irreplaceable. Luckily the prediction tools are better for MHC class I. There is some more limited Class II expression in brain, on microglia and perhaps a subset of neurons, so it is worth looking at too. But overall, class II is expressed on fewer cells and some of those cells are regenerating anyway. It matters where your gene therapy is delivered, though. If it gets into dendritic cells then those will prime a response that spreads to other cell types. The same is true to a lesser extent of monocytes. It is controversial whether or not neutrophils can start an immune response. It would be worth checking if your therapy can get into neutrophil, monocyte, and dendritic.
GFP is very low immunogenic, so you can get long term GFP expression in mice with no immune response. But there are plenty of proteins that if you express them in cells and transplant them into mice, the cells are rejected.
If you really want to study the human immune responses to your gene therapy in mice, both Taconic and JAX actually sell mice that have already been reconstituted with the most common human HLA alleles. You can use these to look for T cell responses based on human HLAs. The animals have murine B and T cells but they adapt to the human HLA. It is also an option to inject human T cells from human blood into mice and study their responses, but this system is more laborious.
